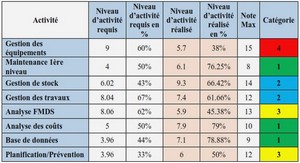
Télécharger le fichier original (Mémoire de fin d’études)
Deinococcus radiodurans
Overview
Deinococcus radiodurans (D. radiodurans) was discovered in 1956 by Arthur W. Anderson (Anderson et al. 1956) during an attempt to sterilize a can of meat using high doses of gamma radiation, supposed to kill all known forms of life. However, after the experiment the meat spoiled. The researchers were able to isolate the bacterium responsible for the spoil and named it Micrococcus radiodurans, later renamed as Deinococcus radiodurans.
D. radiodurans has been found in very different places, ranging from meat to sawdust or in the air of clean rooms and in clothes (Slade & Radman 2011). D. radiodurans was the first Deinococcus isolated, now over 40 of these species have been discovered, in habitats such as hot springs, deserts or Antarctica (De Groot et al. 2005; Ferreira et al. 1997; Hirsch et al. 2004). Among these different species, some present peculiar characteristics directly related to their growth environments (thermophilia, cryophilia) and all present a high resistance to DNA damaging agents.
D. radiodurans is a non-pathogenic, pink-pigmented, 1-3 µm diameter bacterium (Figure 1.1 A), with an outstanding ability to withstand the lethal effects of DNA-damaging events, including ionizing radiation, UV light, and desiccation (Blasius et al. 2008). For example, D. radiodurans can survive, without any loss of viability, UV doses up to 500 J.m-2, a dose that is known to generate ~5,000 pyrimidine dimers per genome copy. It can also resist an acute dose of 5 kGy – irradiation, generating approximately 200 DNA double strand breaks, 3,000 DNA single strand breaks and more than 1,000 damaged bases per genome, a dose representing around 1000 times the maximum dose humans can withstand. D. radiodurans is able to reconstruct a functional genome from hundreds of chromosomal fragments induced by exposure to ionizing radiation, whereas the genomes of most organisms are irreversibly shattered under the same conditions (Figure 1.1 B; reviewed in: (Blasius et al. 2008; Makarova et al. 2001; Slade & Radman 2011)). However, D. radiodurans is not an extremophilic species per se, as it does not thrive in damaging conditions: it can recover and then replicate in normal growth conditions after being exposed to extreme conditions. For example, 85% of D. radiodurans cells survived a 2-year dehydration phase in a desiccator kept below 5% humidity. The ability of D. radiodurans to survive long periods of desiccation may actually be its main characteristic, from which the other resistances could have derived (Mattimore & Battista 1996). Cells that have been engineered to impair their radio-resistance were also shown to be more sensitive to desiccation. All of these unusual properties of D. radiodurans have lead it to be considered as a potential agent for radiodurans post 6.8kGy γ-irradiation. The genome is fully shattered directly after irradiation, as shown by the low molecular weight, smeared bands a t=0, and fully restored in just 3 hours. Adapted from (Rothfuss et al. 2006; Slade & Radman 2011).
The high resistance of D. radiodurans compared to other bacteria (Figure 1.2) has been hypothesized to be due to (i) its ability to protect its proteome, (ii) the efficient repair of its highly shattered genomic information thanks to its protected proteome, (iii) its capacity to export the damaged DNA to the exterior medium.
(i) It has been proposed that high levels of intracellular antioxidant metabolites protect the proteome from reactive oxygen species or free radicals that are produced during irradiation (Daly 2009; Slade & Radman 2011). Even though their mechanisms of action are not really understood, D. radiodurans actually possesses various of these antioxidant metabolites and enzymes, such as Mn- or Fe-superoxide dismutases, Cu/Zn-superoxide dismutases, catalases, peroxidases (Makarova et al. 2001; Wang & Schellhorn 1995), nonenzymic Mn(II) complex (Daly 2009) and carotenoids (Tian et al. 2007). Moreover, D. radiodurans is able to shield its different enzymes from oxidative stress thanks to divalent Mn ions (Mn2+), in complex with small metabolites (such as peptides) (Sharma et al. 2017). For example, Daly et al. showed that a complex, called Mn2+-decapeptide complex (designed according to compounds found in D. radiodurans ultrafiltrates), when injected into mice exposed to 9.5 Gy radiation doses, lead to the 30 days survival of all the mice, whereas only 37% of control mice survived (Gupta et al. 2016).
(ii) Multiple, robust and efficient DNA repair machineries have been observed in D. radiodurans (Daly & Minton 1996; Slade et al. 2009; Zahradka et al. 2006). The different studies of the DNA repair pathways have showed that no “super-proteins” could account for D. radiodurans radioresistance. But, these repair enzymes, although they are well conserved, display numerous differences in structure and/or enzymatic activity compared to their homologues from radio-sensitive bacteria, as reviewed in (Timmins & Moe 2016). These small differences plus the unusual number of DNA glycosylases may add up and significantly contribute to the efficient repair system. The effectiveness of the repair machineries, coupled to the effective protection of the proteome may be key to the resistance of D. radiodurans. Indeed, D. radiodurans can proceed to the repair of its genome thanks to the well protected, and thus functional and efficient, DNA repair proteins.
(iii) The export of the damaged DNA to the exterior medium has also been proposed as a key factor of D. radiodurans resistance. This expulsion would prevent the reinsertion of damaged bases into the genome (Battista 1997). This export may be mediated by UvrA proteins, as they have a close evolutionary relationship to ABC transporter proteins (Doolittle et al. 1986; Linton & Higgins 1998). In particular, UvrA2 which does not appear to play a direct role in the nucleotide excision repair pathway in D. radiodurans may be involved in the export process (White et al. 1999).
Figure 1.2: Difference of survival capacities of several bacteria. Comparison between different species with similar DNA repair protein repertoire, for different levels of irradiation. Adapted from (Daly 2009).
Other factors contribute to its robust phenotype. Its genome is composed of 4 to up to 10 copies of its different chromosomes, varying during the cell cycle (Battista 1997). This high redundancy of genetic information may therefore ensure the presence, and the availability, of intact DNA template for the restoration of the original genome (Minton & Daly 1995). Moreover, its genome is particularly condensed, which is proposed to prevent the dispersion of free DNA ends (Minsky et al. 2006). Interestingly, D. radiodurans genome has been shown to adopt a ring-like structure that is unaffected by high doses of gamma irradiation (Englander et al. 2004; Levin-Zaidman et al. 2003). Condensed genomes appear to be a common feature of a number of radiation resistant bacteria (Zimmerman & Battista 2005). The link between nucleoid organisation andrans bacterial radioresistance is further discussed in “2.3 D. radiodurans chromosome organization and condensation”.
In addition to this peculiar phenotype, D. radiodurans also possesses numerous properties making it suitable for lab work: D. radiodurans is a class 1 organism, it can be cultivated in the laboratory in rich medium and a wide range of genetic tools has been developed over the years in particular in Prof. Sommer’s laboratory in Orsay (now directed by Prof. Confalonieri), with whom we collaborated for the work presented in this manuscript (Lecointe et al. 2004).
Table des matières
Table of Contents
Table of Figures
Table of Tables
Table of Abbreviations and Acronyms
Chapter I: Introduction
1. Deinococcus radiodurans
1.1. Overview
1.2. Cell wall structure
2. Nucleoid organization
2.1. Molecular organization by nucleoid-associated proteins
2.2. Micro- & macro-domain organization
2.3. Spatial organization of the nucleoid
2.4. Nucleoid compaction mechanisms
2.5. D. radiodurans chromosome organization and condensation
3. Chromosome segregation
3.1. ParABS system, an active model for origin segregation
3.2. Passive model for bulk chromosome segregation
3.3. Segregation termination
3.4. Segregation in D. radiodurans
4. Bacterial cell division
4.1. Rod vs. (ovo)cocci division
4.2. Division site selection and cytokinesis
4.3. D. radiodurans cell division
Chapter II: Overview of fluorescence microscopy
1. Optical microscopy
2. History of fluorescence microscopy
3. Fluorescence
4. Fluorophores
5. Optical resolution principle
6. Conventional fluorescence microscopy
7. SR fluorescence microscopy
7.1. Ensemble
7.2. Single-molecule
Chapter III: Objectives of the PhD
1. Autoblinking: a SR artifact
2. D. radiodurans cell and nucleoid dynamics
3. Dissemination
Chapter IV: Autoblinking
Chapter V: D. radiodurans cell and nucleoid dynamics
1. Abstract
2. Introduction
3. Results
4. Discussion
5. Methods
6. References
7. Supplementary material
Chapter VI: Discussion and perspectives
1. Autoblinking
2. D. radiodurans cell and nucleoid dynamics
2.1. Cell morphology definition
2.2. Cell growth
2.3. Nucleoid organization, segregation and compaction
2.4. Perspectives
3. D. radiodurans imaging
Chapter VII: Appendices
1. Codes used for analyses
1.1. Density graph of foci
1.2. Perimeter fraction of a phase 1 diad
1.3. Translation of trackmate output to trackart input
2. Video material
3. Résumé de la thèse en Français
Bibliography
Acknowledgment
Resumé
Summary
Télécharger le rapport complet